Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T83369
(Former ID: TTDC00078)
|
|||||
| Target Name |
Coagulation factor IX (F9)
|
|||||
| Synonyms |
Plasma thromboplastin component; PTC protein; Factor IX; Christmas factor
Click to Show/Hide
|
|||||
| Gene Name |
F9
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 2 Target-related Diseases | + | ||||
| 1 | Bleeding disorder [ICD-11: GA20-GA21] | |||||
| 2 | Coagulation defect [ICD-11: 3B10] | |||||
| Function |
Factor IX is a vitamin K-dependent plasma protein that participates in the intrinsic pathway of blood coagulation by converting factor X to its active form in the presence of Ca(2+) ions, phospholipids, and factor VIIIa.
Click to Show/Hide
|
|||||
| BioChemical Class |
Peptidase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 3.4.21.22
|
|||||
| Sequence |
MQRVNMIMAESPGLITICLLGYLLSAECTVFLDHENANKILNRPKRYNSGKLEEFVQGNL
ERECMEEKCSFEEAREVFENTERTTEFWKQYVDGDQCESNPCLNGGSCKDDINSYECWCP FGFEGKNCELDVTCNIKNGRCEQFCKNSADNKVVCSCTEGYRLAENQKSCEPAVPFPCGR VSVSQTSKLTRAETVFPDVDYVNSTEAETILDNITQSTQSFNDFTRVVGGEDAKPGQFPW QVVLNGKVDAFCGGSIVNEKWIVTAAHCVETGVKITVVAGEHNIEETEHTEQKRNVIRII PHHNYNAAINKYNHDIALLELDEPLVLNSYVTPICIADKEYTNIFLKFGSGYVSGWGRVF HKGRSALVLQYLRVPLVDRATCLRSTKFTIYNNMFCAGFHEGGRDSCQGDSGGPHVTEVE GTSFLTGIISWGEECAMKGKYGIYTKVSRYVNWIKEKTKLT Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 2 Approved Drugs | + | ||||
| 1 | Emicizumab | Drug Info | Approved | Factor VIII deficiency | [2] | |
| 2 | Recombinant Factor IX | Drug Info | Approved | Bleeding disorder | [3] | |
| Clinical Trial Drug(s) | [+] 5 Clinical Trial Drugs | + | ||||
| 1 | CSL-654 | Drug Info | Phase 3 | Factor IX deficiency | [7] | |
| 2 | RG6013 | Drug Info | Phase 3 | Haemophilia A | [8] | |
| 3 | AMT-060 | Drug Info | Phase 1/2 | Factor IX deficiency | [11] | |
| 4 | AAV2-hFIX16 | Drug Info | Phase 1 | Factor IX deficiency | [13] | |
| 5 | N9-GP | Drug Info | Phase 1 | Factor IX deficiency | [14] | |
| Discontinued Drug(s) | [+] 4 Discontinued Drugs | + | ||||
| 1 | RB-006 | Drug Info | Discontinued in Phase 3 | Vascular disease | [15] | |
| 2 | Trenonacog alfa | Drug Info | Discontinued in Phase 3 | Discovery agent | [16] | |
| 3 | SB 249417 | Drug Info | Discontinued in Phase 1 | Sepsis | [17] | |
| 4 | Draculin | Drug Info | Terminated | Thrombosis | [18] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Modulator | [+] 11 Modulator drugs | + | ||||
| 1 | Emicizumab | Drug Info | [2] | |||
| 2 | Recombinant Factor IX | Drug Info | [1] | |||
| 3 | CSL-654 | Drug Info | [19] | |||
| 4 | AMT-060 | Drug Info | [21] | |||
| 5 | AAV2-hFIX16 | Drug Info | [22] | |||
| 6 | N9-GP | Drug Info | [23] | |||
| 7 | Trenonacog alfa | Drug Info | [16] | |||
| 8 | Draculin | Drug Info | [26] | |||
| 9 | F-9TG | Drug Info | [28] | |||
| 10 | Factor IX-XTEN | Drug Info | [28] | |||
| 11 | MOD-3012 | Drug Info | [28] | |||
| Inhibitor | [+] 7 Inhibitor drugs | + | ||||
| 1 | RB-006 | Drug Info | [24] | |||
| 2 | 4-iodobenzo[b]thiophene 2-carboxamidine | Drug Info | [27] | |||
| 3 | 4-Phenyl-benzo[b]thiophene-2-carboxamidine | Drug Info | [27] | |||
| 4 | 6-Benzyloxybenzo[b]thiophene-2-carboxamidine | Drug Info | [27] | |||
| 5 | Gamma-Carboxy-Glutamic Acid | Drug Info | [29] | |||
| 6 | PMID20121197C57 | Drug Info | [30] | |||
| 7 | RAZAXABAN | Drug Info | [31] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: 2-Methyl-2-Propanol | Ligand Info | |||||
| Structure Description | HUMAN COAGULATION FACTOR IXA IN COMPLEX WITH P-AMINO BENZAMIDINE | PDB:1RFN | ||||
| Method | X-ray diffraction | Resolution | 2.80 Å | Mutation | No | [32] |
| PDB Sequence |
VVGGEDAKPG
25 QFPWQVVLNG35 KVDAFCGGSI46 VNEKWIVTAA56 HCVETGVKIT65 VVAGEHNIEE 75 TEHTEQKRNV85 IRIIPHHNYN95 AAINKYNHDI103 ALLELDEPLV113 LNSYVTPICI 123 ADKEYTNIFL131 KFGSGYVSGW141 GRVFHKGRSA152 LVLQYLRVPL162 VDRATCLRST 172 KFTIYNNMFC182 AGFHEGGRDS190 CQGDSGGPHV200 TEVEGTSFLT210 GIISWGEECA 221 MKGKYGIYTK230 VSRYVNWIKE240 KTKLT
|
|||||
|
|
||||||
| Ligand Name: UDP-glucose | Ligand Info | |||||
| Structure Description | Crystal structure of mouse Xyloside xylosyltransferase 1 complexed with manganese,acceptor ligand and UDP-Glucose | PDB:4WMA | ||||
| Method | X-ray diffraction | Resolution | 1.62 Å | Mutation | No | [33] |
| PDB Sequence |
QCESNPCLNG
59 GSCKDDINSY69 ECWCPFGFEG79 KNCEL
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Complement and coagulation cascades | hsa04610 | Affiliated Target |
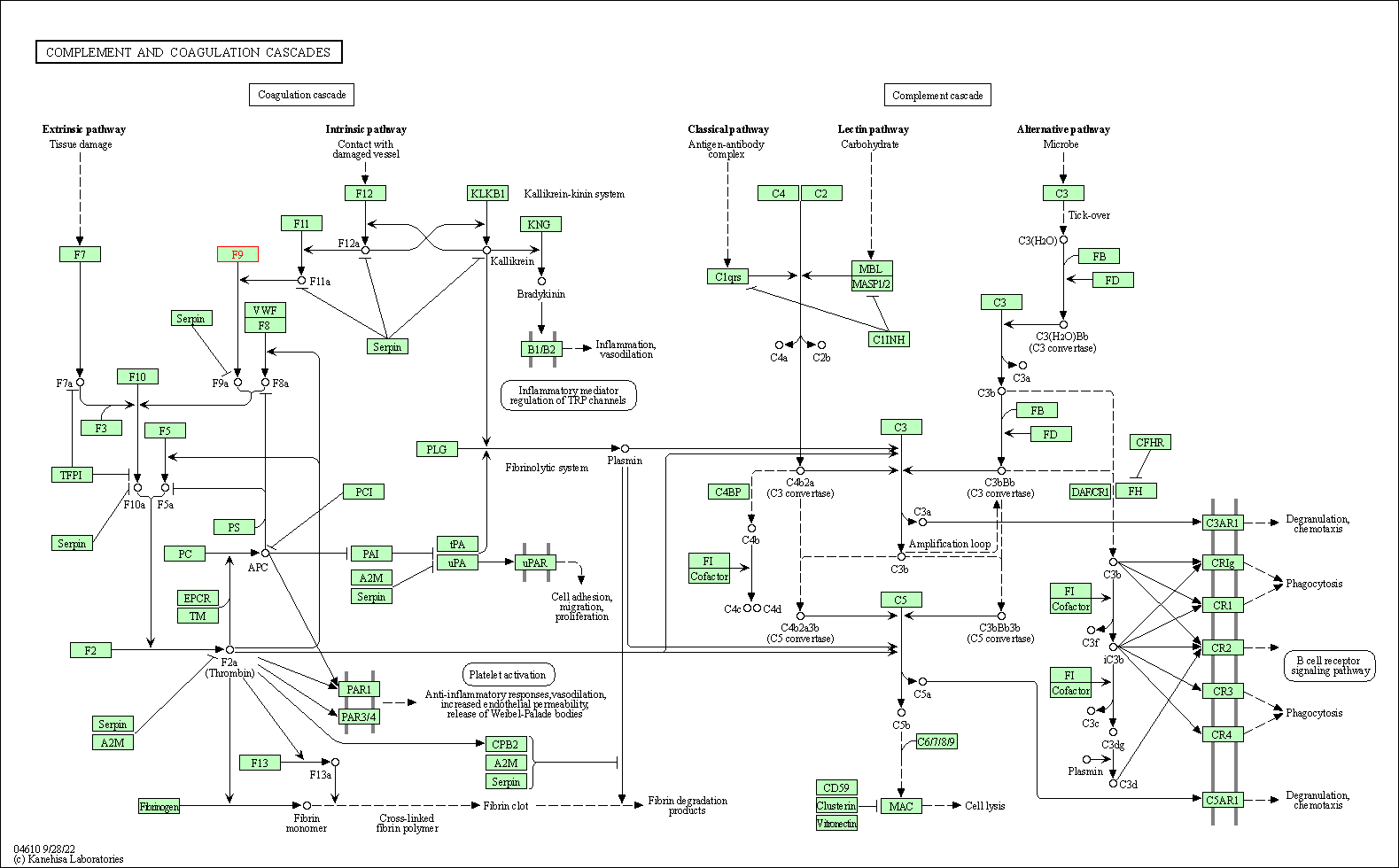
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Degree | 7 | Degree centrality | 7.52E-04 | Betweenness centrality | 2.08E-05 |
|---|---|---|---|---|---|
| Closeness centrality | 1.80E-01 | Radiality | 1.30E+01 | Clustering coefficient | 4.29E-01 |
| Neighborhood connectivity | 1.06E+01 | Topological coefficient | 2.52E-01 | Eccentricity | 13 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating Transcription Factors | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 1 KEGG Pathways | + | ||||
| 1 | Complement and coagulation cascades | |||||
| Panther Pathway | [+] 1 Panther Pathways | + | ||||
| 1 | Blood coagulation | |||||
| Pathwhiz Pathway | [+] 1 Pathwhiz Pathways | + | ||||
| 1 | Coagulation | |||||
| Reactome | [+] 5 Reactome Pathways | + | ||||
| 1 | Extrinsic Pathway of Fibrin Clot Formation | |||||
| 2 | Intrinsic Pathway of Fibrin Clot Formation | |||||
| 3 | Gamma-carboxylation of protein precursors | |||||
| 4 | Transport of gamma-carboxylated protein precursors from the endoplasmic reticulum to the Golgi apparatus | |||||
| 5 | Removal of aminoterminal propeptides from gamma-carboxylated proteins | |||||
| WikiPathways | [+] 4 WikiPathways | + | ||||
| 1 | Complement and Coagulation Cascades | |||||
| 2 | PTM: gamma carboxylation, hypusine formation and arylsulfatase activation | |||||
| 3 | Blood Clotting Cascade | |||||
| 4 | Formation of Fibrin Clot (Clotting Cascade) | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | BAX326 (RIXUBIS): a novel recombinant factor IX for the control and prevention of bleeding episodes in adults and children with hemophilia B. Ther Adv Hematol. 2014 Oct;5(5):168-80. | |||||
| REF 2 | 2017 FDA drug approvals.Nat Rev Drug Discov. 2018 Feb;17(2):81-85. | |||||
| REF 3 | DAILYMED.nlm.nih.gov:IXINITY- coagulation factor ix (recombinant) | |||||
| REF 4 | ClinicalTrials.gov (NCT03569891) HOPE-B: Trial of AMT-061 in Severe or Moderately Severe Hemophilia B Patients. U.S. National Institutes of Health. | |||||
| REF 5 | ClinicalTrials.gov (NCT03861273) A Study to Evaluate the Efficacy and Safety of Factor IX Gene Therapy With PF-06838435 in Adult Males With Moderately Severe to Severe Hemophilia B (BENEGENE-2). U.S. National Institutes of Health. | |||||
| REF 6 | ClinicalTrials.gov (NCT05878938) Open-label Safety Study in Adults and Adolescents With Haemophilia A With and Without FVIII Inhibitors Switching Directly From Emicizumab Prophylaxis to NNC0365-3769 (Mim8) Prophylaxis. U.S.National Institutes of Health. | |||||
| REF 7 | ClinicalTrials.gov (NCT02053792) A Safety and Efficacy Extension Study of a Recombinant Fusion Protein Linking Coagulation Factor IX With Albumin (rIX-FP) in Patients With Hemophilia B. U.S. NationalInstitutes of Health. | |||||
| REF 8 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 9 | Partial factor IXa inhibition with TTP889 for prevention of venous thromboembolism: an exploratory study. J Thromb Haemost. 2008 Mar;6(3):457-63. | |||||
| REF 10 | ClinicalTrials.gov (NCT02695160) Ascending Dose Study of Genome Editing by Zinc Finger Nuclease Therapeutic SB-FIX in Subjects With Severe Hemophilia B. U.S. National Institutes of Health. | |||||
| REF 11 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800026919) | |||||
| REF 12 | Clinical pipeline report, company report or official report of Roche | |||||
| REF 13 | ClinicalTrials.gov (NCT00515710) LTFU for Gene Transfer Subjects With Hemophilia B. U.S. National Institutes of Health. | |||||
| REF 14 | ClinicalTrials.gov (NCT01395810) Safety and Efficacy of NNC-0156-0000-0009 After Long-Term Exposure in Patients With Haemophilia B: An Extension to Trials NN7999-3747 and NN7999-3773. U.S. National Institutes of Health. | |||||
| REF 15 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800019932) | |||||
| REF 16 | Pharmacokinetic properties of IB1001, an investigational recombinant factor IX, in patients with haemophilia B: repeat pharmacokinetic evaluation and sialylation analysis.Haemophilia.2012 Nov;18(6):881-7. | |||||
| REF 17 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800010912) | |||||
| REF 18 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800004668) | |||||
| REF 19 | Safety and pharmacokinetics of a novel recombinant fusion protein linking coagulation factor IX with albumin (rIX-FP) in hemophilia B patients. Blood. 2012 September 20; 120(12): 2405-2411. | |||||
| REF 20 | Anti-factor IXa/X bispecific antibody (ACE910): hemostatic potency against ongoing bleeds in a hemophilia A model and the possibility of routine supplementation. J Thromb Haemost. 2014 Feb;12(2):206-13. | |||||
| REF 21 | Phase I/II clinical trial of AMT-060 for treating hemophilia B. uniQure N.V. | |||||
| REF 22 | Assessing the potential for AAV vector genotoxicity in a murine model. Correction in: Blood. 2011 June 16; 117(24): 6739. | |||||
| REF 23 | Enhanced pharmacokinetic properties of a glycoPEGylated recombinant factor IX: a first human dose trial in patients with hemophilia B. Blood. 2011 Sep 8;118(10):2695-701. | |||||
| REF 24 | In-vitro evaluation of anti-factor IXa aptamer on thrombin generation, clotting time, and viscoelastometry. Thromb Haemost. 2009 May;101(5):827-33. | |||||
| REF 25 | Pharmacokinetic and pharmacodynamic modeling of humanized anti-factor IX antibody (SB 249417) in humans. Clin Pharmacol Ther. 2002 Apr;71(4):235-45. | |||||
| REF 26 | Expression of biological activity of draculin, the anticoagulant factor from vampire bat saliva, is strictly dependent on the appropriate glycosylation of the native molecule. Biochim Biophys Acta. 1998 Oct 23;1425(2):291-9. | |||||
| REF 27 | Studies of benzothiophene template as potent factor IXa (FIXa) inhibitors in thrombosis. J Med Chem. 2010 Feb 25;53(4):1465-72. | |||||
| REF 28 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 2364). | |||||
| REF 29 | How many drug targets are there Nat Rev Drug Discov. 2006 Dec;5(12):993-6. | |||||
| REF 30 | Structure based drug design: development of potent and selective factor IXa (FIXa) inhibitors. J Med Chem. 2010 Feb 25;53(4):1473-82. | |||||
| REF 31 | Aminobenzisoxazoles with biaryl P4 moieties as potent, selective, and orally bioavailable factor Xa inhibitors. Bioorg Med Chem Lett. 2006 Apr 1;16(7):1795-8. | |||||
| REF 32 | Coagulation factor IXa: the relaxed conformation of Tyr99 blocks substrate binding. Structure. 1999 Aug 15;7(8):989-96. | |||||
| REF 33 | Notch-modifying xylosyltransferase structures support an SNi-like retaining mechanism. Nat Chem Biol. 2015 Nov;11(11):847-54. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

